Benchmarking molecular fingerprints
Inspired by Anthony Nicholls's paper "So what do we know and when do we know it?", I've started measuring the performance of available molecular fingerprint methods in discriminating between active and inactive componuds. Here are some preliminary results.
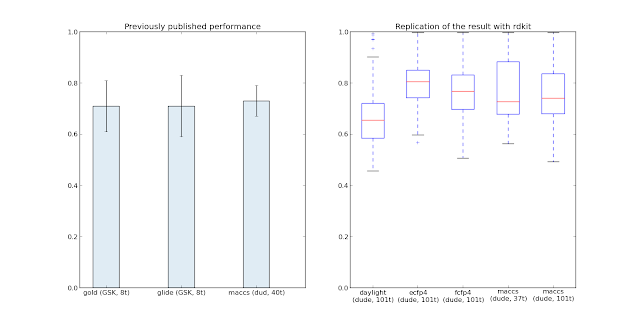
The left side of the first figure summarizes published results in the field; the right side of the figure reproduces the known performance for MACCS fingerprint and adds-in a bunch of different fingerprints. Overall they're not that different with ECFP4 perhaps being the best (highest median and narrow 1st-Quartile-3rd-Quartile range).
All methods were tested on the same ~100 protein targets. The second figure tries to explain how the performance of various fingerprints may be correlated with each other across this set of 100 targets.
The left side of the first figure summarizes published results in the field; the right side of the figure reproduces the known performance for MACCS fingerprint and adds-in a bunch of different fingerprints. Overall they're not that different with ECFP4 perhaps being the best (highest median and narrow 1st-Quartile-3rd-Quartile range).

Comments
Post a Comment