Deposit Silicos-it Shape-it as an Ubuntu 12.04 package in a personal PPA
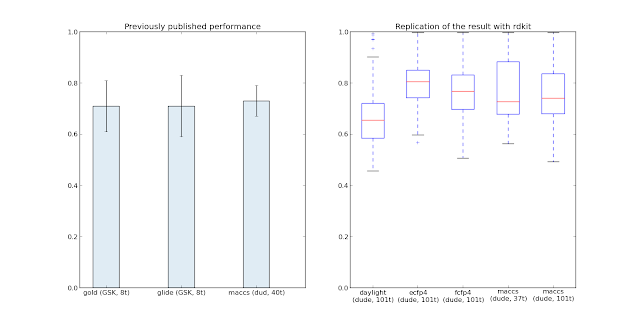
See, I've never made a package for ubuntu repos, so I didn't know how much fun I was in... Shape-it an open and free software package that's using the same underlying theory as OpenEye's ROCS. Both codes perform a Gaussian-overlap of groups of atoms. But free stuff is good and so to make the free stuff even more free I've decided to put it in the Ubuntu repos. Now, I don't know how the Silicios-it and OpenEye implementations compare in either speed or getting the "right" answer (data for getting the right answer is know for ROCS from benchmarks on the DUD set, for example), so no guarantees folks! Piecing together bits from various tutorials, I've come up with the package.sh script below that will download the original source and configure the package for you. Replace jandom-gmail and other details with your data, setup a PPA on launchpad, generate a GPG public-private keypair and register it with the PPA - you should be ready to go. ...