Measuring the performance of ligand-based methods
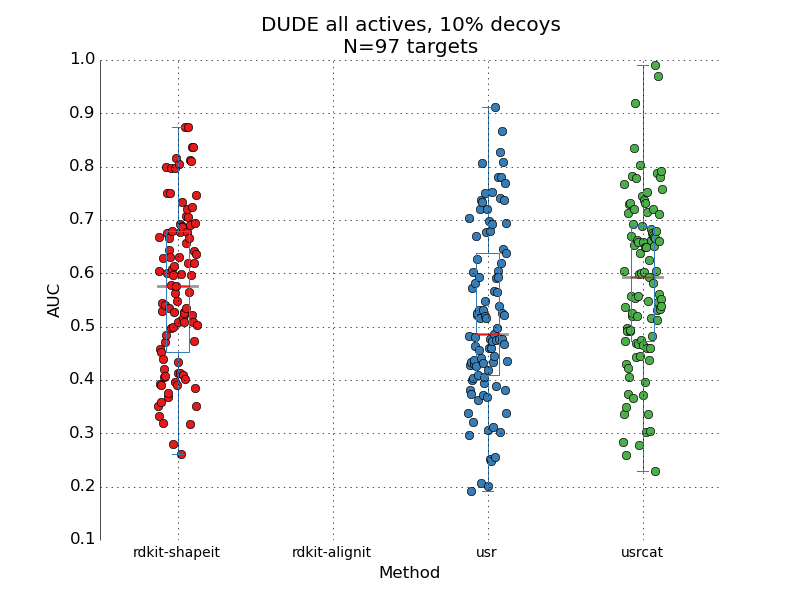
This is another episode in the "how good are these methods really?" series. The aim is to understand how well the ligand-based 3D methods for virtual screening perform under standard benchmarking sets. In the original papers introducing the methods frequently is benchmarking done on a small test sets, prone to all sorts of small population problems. Here I take 3 popular methods: the ROCS-like implementation of Gaussian shape overlap[1] that I recently ported to rdkit as well as USR[2] and USR-CAT[3], two very fast shape methods. All 3 are tested on the DUDE[4] dataset, a standard benchmarking set. Conformer generation was done as outlined by JP in his paper[5]. Pair of methods compared Statistic used Value rdkit-shape VS usr wilcoxon T=2.24, p=0.02 rdkit-shape VS usrcat wilcoxon T=-0.47, p=0.63 usr VS usrcat wilcoxon T=-2.70, p=0.006 One thing that struck me was the spread of performance. I got a little worried that the conformer ...